Fetal Health Classification
Classify fetal health to prevent child and maternal mortality
1 Abstract
The goal of this study is to classify fetal health in 3 categories in order to prevent child and maternal mortality (Normal, Suspicious and Pathological). By testing medicale cardiotocographic data ( from Kaggle) on 3 models (KNN, LinearSVC,C-SVC), the more accurate model to predict the health criticity of the patients is the KNN model. The PCA modelisation and correlation prove that all CTG variables are need to achieve a good prediction.
2 Motivation
In order to achieve the UNICEF Sustainable Development Goals, all countries aiming to reduce under‑5 mortality to at least as low as 25 per 1,000 live births. Cardiotocograms (CTGs) are a simple and cost accessible medical devices to assess fetal health. With ultrasound the CTG measure fetal heart rate, fetal movements, uterine contractions…(11 measurements). A good indicator of the fetal health is valuable for healthcare professionals.
3 Dataset
The data set is a csv file downloaded from Kaggle website. It is compound of 2126 samples and 22 variables. Data came from Ayres de Campos et al. (2000) SisPorto 2.0 A Program for Automated Analysis of Cardiotocograms. J Matern Fetal Med 5:311-318.
4 Data preparation and Cleaning
The most important preparation is to check for the absolute absence of patient identity. With the help of a midwife we select the variable available in most CTG. The study will contain 2016 samples and 11 variables.
Variables
baseline value, accelerations, fetal_movement, uterine_contractions, light_decelerations, severe_decelerations , prolongued_decelerations , Abnormal_short_term_variability, mean_short_term_variability, abnormal_long_term_variability, long_term_variability.
5 Research Questions
Main question: Is it possible to predict fetal health with CTG variables? With what precision?
Secondary question: When predict fetal health, does some CTG variables useless ? Could we simplify the CTG interface?
6 Methods
Does some CTG variables useless? a/ Correlation between variables b/ Principal Components Analysis (PCA)
Is it possible to predict fetal health? With what precision? a/ Find the best fitting model, data split b/ Support vector machines: LinearSVC c/ Nearest Neighbors Classification d/ Support vector machines: C-Support Vector Classification
Findings: Does some CTG variables more usefull?
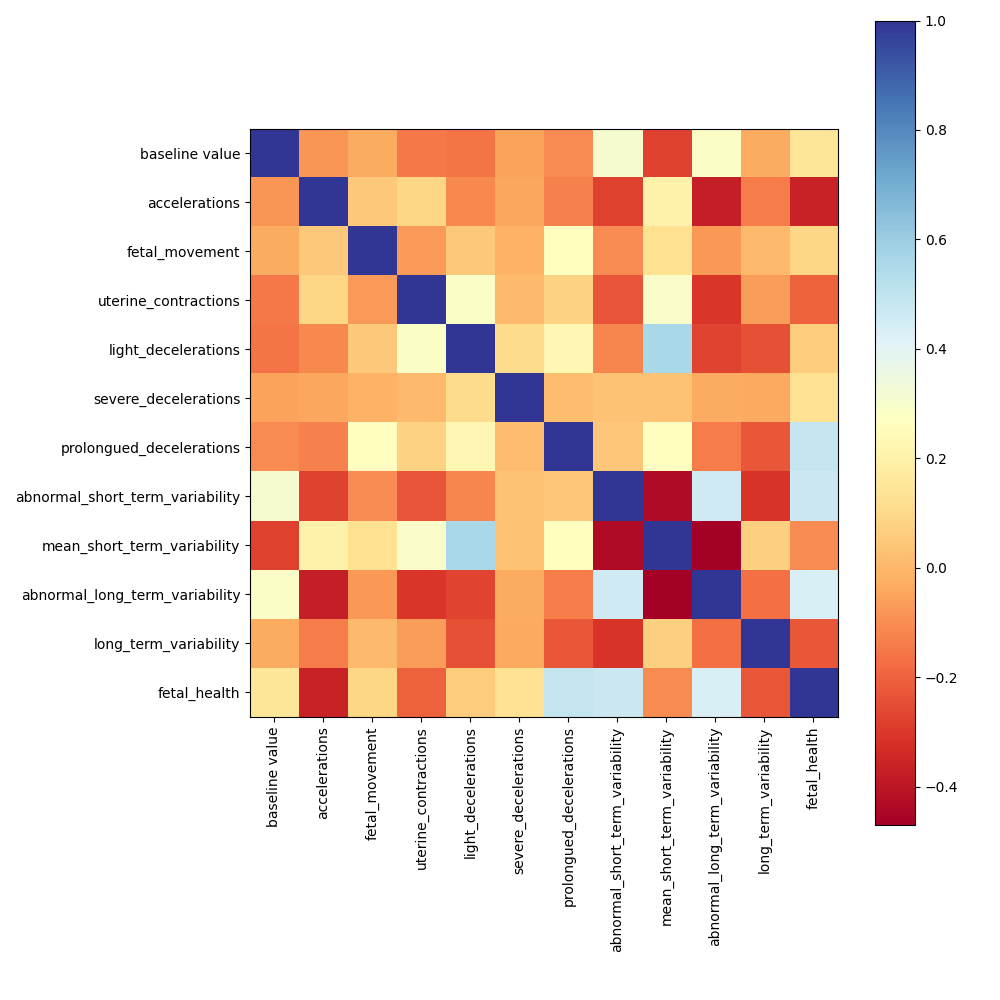
We can't see any strong correlation between the variables (less than 0.5). A variables reduction will be difficult.
Principal component analysis of the samples
Correlation between variables
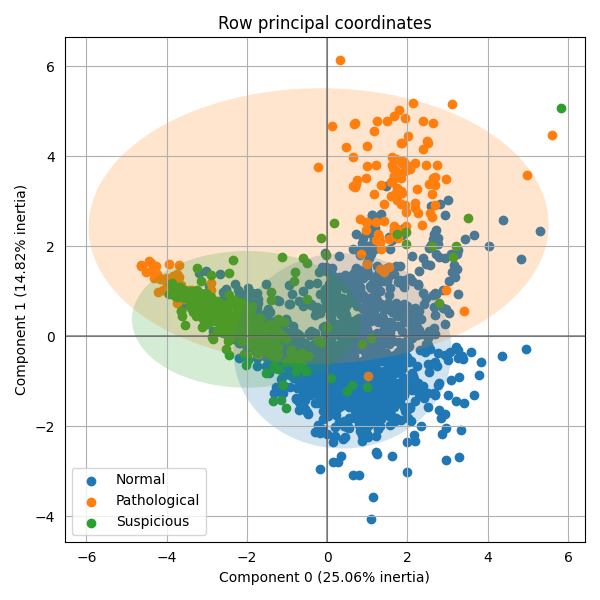
The PCA show us that the 3 groups are mixed. Generally, the more the groups are mixed, the more variables are need to achieve an accurate classification.
Principal component analysis of the variables
The variables PCA shows spread variable around the circle. The vector of most of the variables is long (>0.5). Wich mean that the variable reduction won’t be possible without losing too much information.
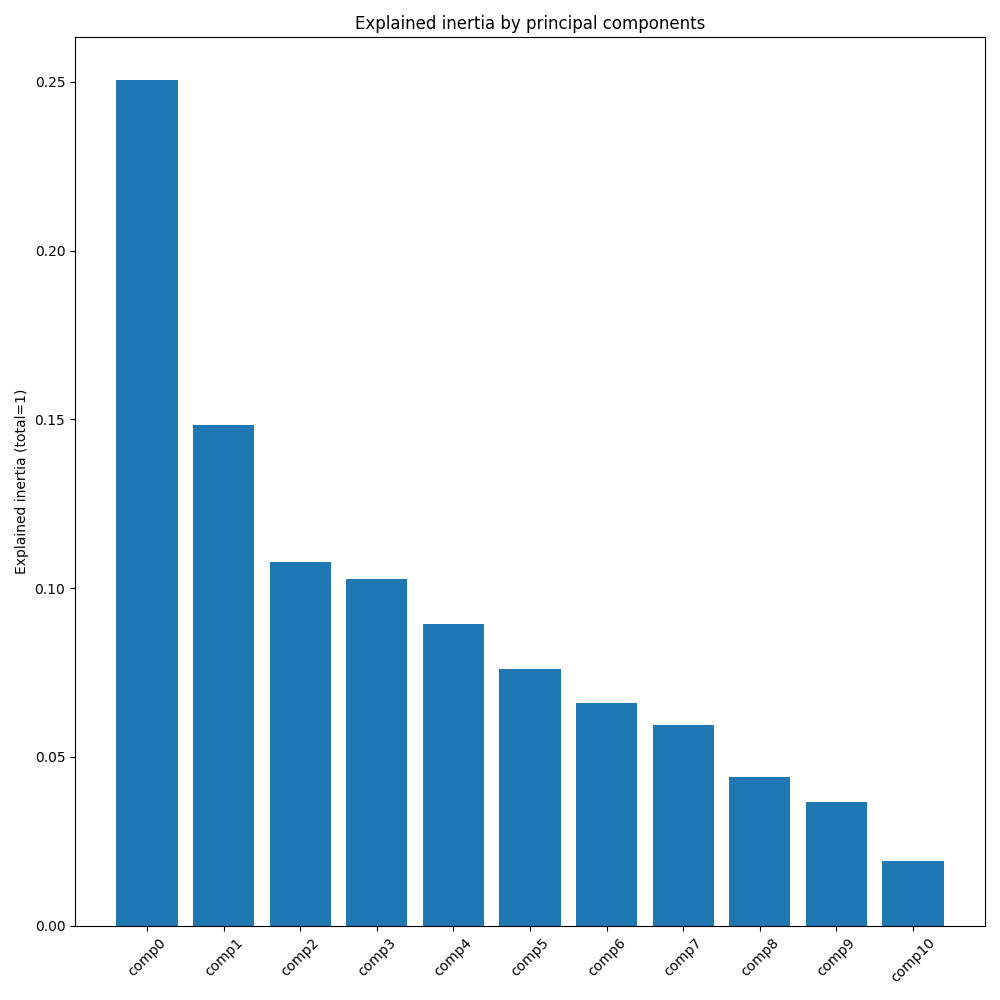
Explained inertia by principal components

The main component (0) is small: 0.25. The second too: 0.15 Wich mean that these 2 components explain only 40% of the information. The total of the other component explain 60% of the information. To explain a fetal health accurately the 11 variables are needed.
Findings: Is it possible to predict fetal health?
a/Find the best fitting model, data split
The samples had been split in 2 group: 40% as train samples 60% as test samples
10 runs had been done with each model in order to ensure results stability.
Support vector machines (SVMs) LinearSVC
LinearSVC with 10000 iteration => accuracy score: 0.786
Nearest Neighbors Classification
KNeighborsClassifier(n_neighbors=3) => accuracy score: 0.874
Support vector machines: C-Support Vector Classification
SVC() => accuracy score: 0.813
The KNN is the more accurate over the 3 models.
Findings: Test of the selected model with midwives usual values
his is a real life model test using the variables traced by midwives in the patient report: baseline value, accelerations, uterine_contractions, light_decelerations, severe_decelerations, prolongued_decelerations, long_term_variability.
KNeighborsClassifier(n_neighbors=3) => accuracy score: 0.756
Because of the lack of some values, the model accuracy is lower: 0.756 < 0.874
Limitations
Because of the limitation of the number of samples, the model is fragile. With a larger sample, the model would be more accurate and thrustable. Other important variables are missing, like gestational age and pregnancy pathologies.
Conclusions
ACT and correlation between variables prove that all 11 variables are needed for accurate diagnostics. In clinic, the midwives usent all of those variable to estimate the patient risk.
The model use to predict fetal health score an accuracy of more than 85%. The prediction seems good, but only as a proposal value, not for a medical diagnostic.
Acknowledgements
Data set from Ayres de Campos et al. (2000) SisPorto 2.0 A Program for Automated Analysis of Cardiotocograms. J Matern Fetal Med 5:311-318.
Contribution of a midwife from a private clinic center.
References
UNESCO: Child survival and the SDGs (2020).
SisPorto 2.0: A Program for Automated Analysis of Cardiotocograms. J Matern Fetal Med 5:311-318
Code
feel free to use it
By Benoit Pont